Guide for the Generation of Gene Knock-outs by Targeted Deletions or Insertions
This guide illustrates how to make a complete gene knock-out or in other terms, a null-allele.
This can be achieved by deleting parts of the target gene that effect essential functions. In contrast, early frame-shift mutations generated by one sgRNA often result in incomplete knock-outs, or in other terms, hypomorphs [Ref].
To get started, retrieve a gene model and genomic sequence of your knock-out target gene from Ensembl, FlyBase or similar. Protein annotations are below the gene model, for example, PROSITE, Pfam, see link here
For illustration, we use Drosophila as a model to explain the process. A main advantage of this model organism is the availability of transposon inserts at the beginning of many genes. Classic approaches used imprecise excision of P-elements to generate deletions as described above. Here, we design sgRNAs that flank the transposon to then screen for loss of the transposon marker (e.g. GFP) after sgRNA/Cas9 mediated excision in the germline. In principle, this approach can be adapted to other model organisms and use a PCR screening approach to identify positive individuals.
For cell-lines the approach is in principle the same, but relies on chronic exposure of sgRNA/Cas9 (under selection, e.g. Puromycin) and a PCR approach for screening as detailed below.
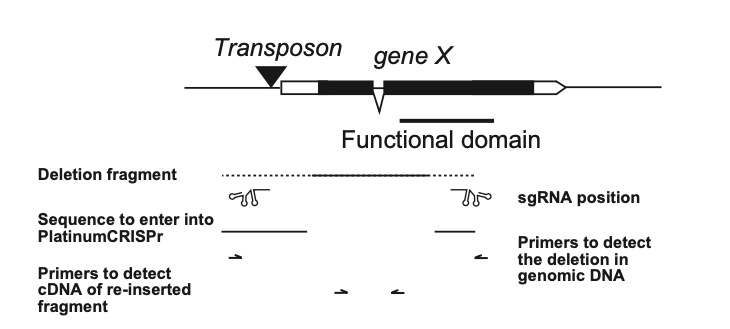
Schematic of a typical Drosophila gene indicating the position of sgRNAs and a deletion to be generated to obtain a knock-out below the gene model. The line indicates the desired deletion and flanking dashed lines indicate positions where gRNAs can be selected. Below are primers indicated to screen for the deletion which will result in a short PCR product if the deletion is present compared to a long PCR product in the absence of a deletion, which will likely not amplify efficiently. Then, to screen for re-insertions, primers are indicated flanking an intron in the deleted part. If re-integration occurs, a short PCR product will be amplified for integration in the sense direction. If a longer PCR product is amplified, indicating a non-spliced intron, integration will be in the anti-sense direction.
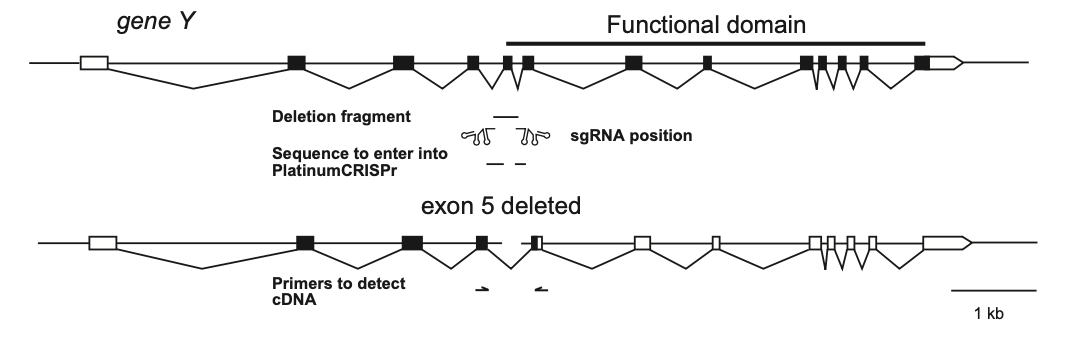
Schematic of a typical human gene composed of short exons and large introns with the translated part marked in black. Removal of an exon can lead to a frameshift resulting in a truncated protein, that is non-functional (below). The line indicates the desired deletion induced by the indicated sgRNAs. RT-PCR with primers in exons flanking the deleted exon can be used to monitor CRISPR-Cas9 induced deletion efficiency in cultured cells based on the ration of splice products.
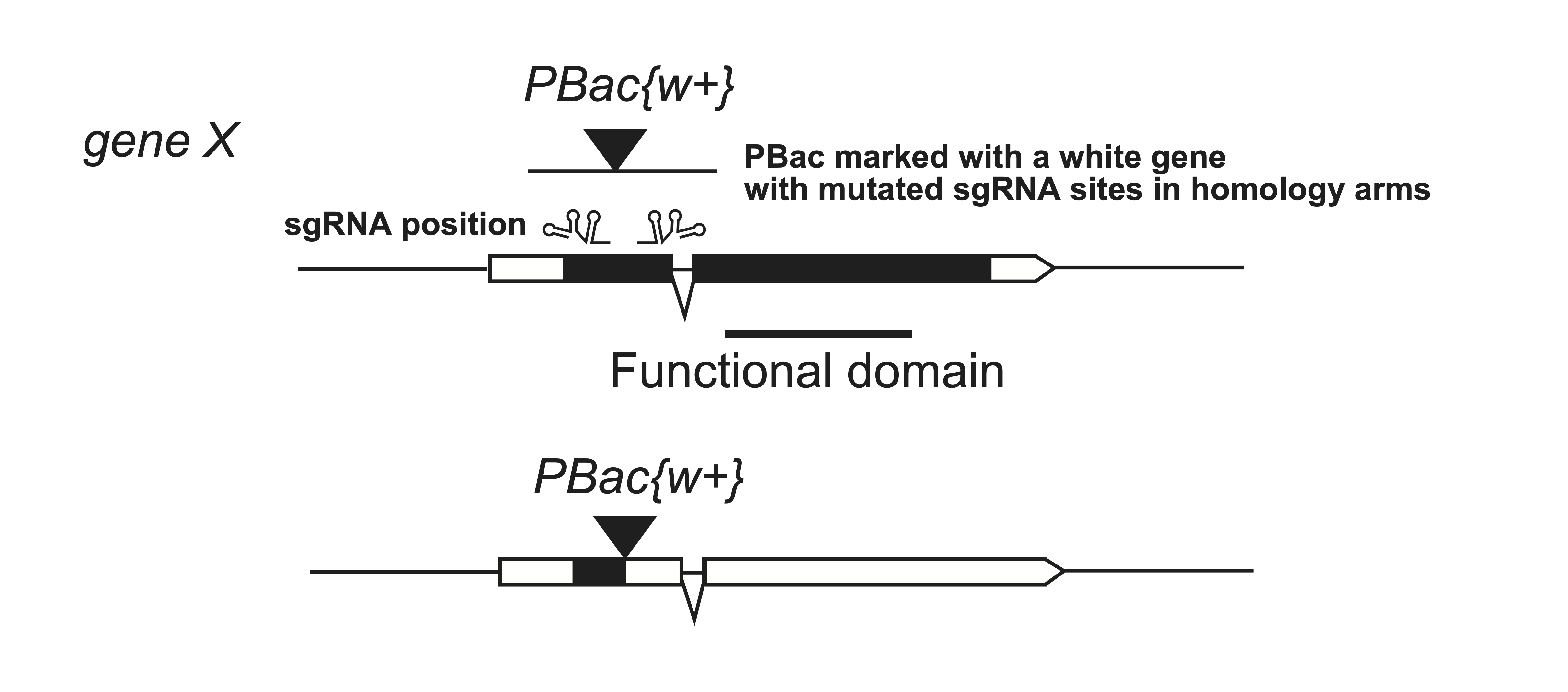
In Drosophila, we have generated a white gene containing PBac vector [Ref], that can be inserted in the beginning of a given gene to generate a truncated protein. Insertions in a white mutant background can then be recognized by a red eye color. If the PBac is inserted at a TTAA sequence, it can later be removed by a PBac transposase. Note, when using homology arm construct for repair, the sgRNA sites in the homology arm need to be mutated to prevent cleavage.
An extension of this approach is to include mutations in the homology arms that affect protein function. These mutations will become effective after excision of the PBac transposon.
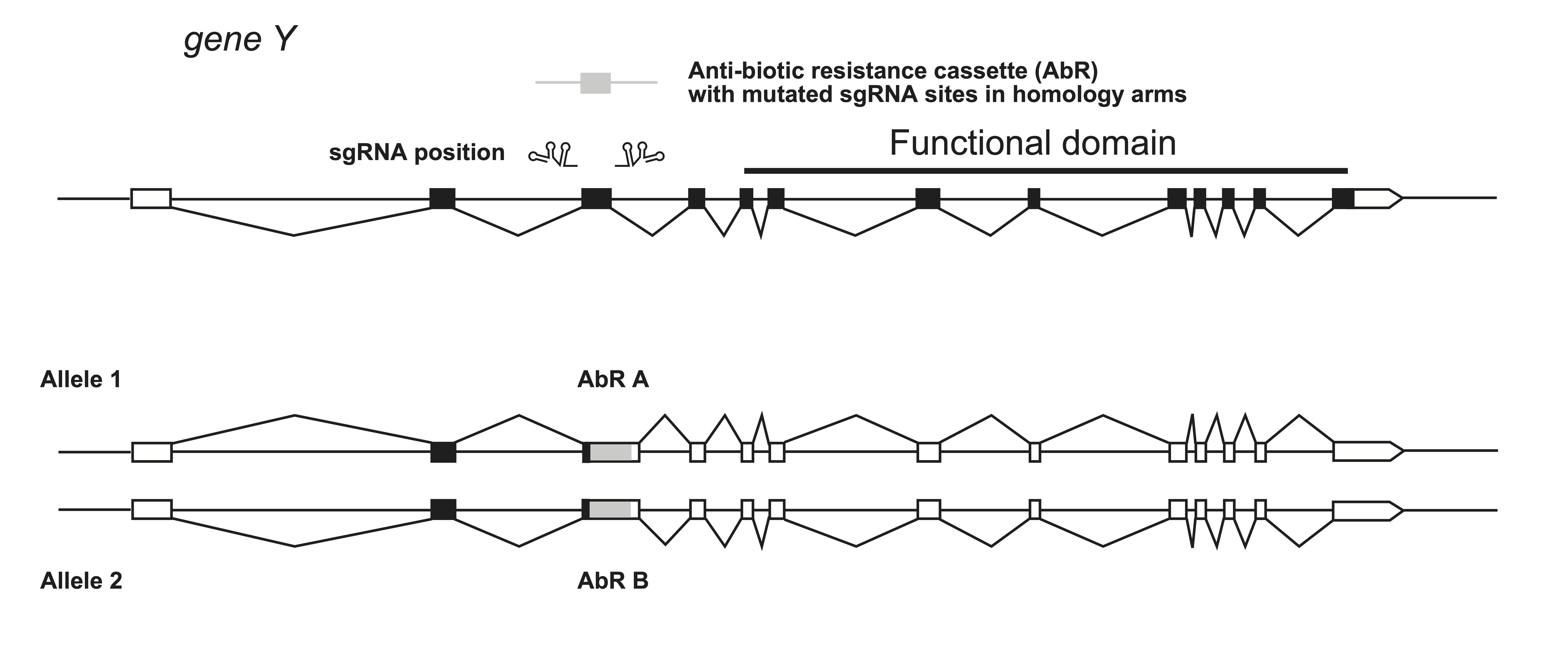
An alternative way to make knock-outs in cultured cells, is to insert an antibiotic selection cassette in the beginning of the gene to result in a truncated protein. To ensure both alleles are knocked-out, a second insertion cassette is used, which contains a different antibiotic selection (cells are efficiently eliminated with 2.5 mg/ml neomycin, 400 µg/ml hygromycin, 0.75 µg/ml puromycin, or 4 µg/ml blasticidin S, Ishibashi et al, 2020.)
To generate conditional knock-out alleles, a degron module can be incorporated at the end of the gene (Nabet et al. 2018) again by using two selection cassettes to target both alleles.
Of note, many cell lines are aneuploid and potentially can contain more than two copies of a given gene. Also, insertion of degron cassette at the end of a gene can effect protein function.
Disclaimer
To get started, retrieve a gene model and genomic sequence of your knock-out target gene from Ensembl, FlyBase or similar. Protein annotations are below the gene model, for example, PROSITE, Pfam, see link here
Gene deletion mutagenesis
An ideal way to a gene knock-out would be to remove a part of the promotor (which is the TATA box), and part of a functional domain in the target gene, for example catalytic domains, DNA binding domains, RNA binding domains or similar. As we have observed that the deleted parts can re-integrate, neither the re-inserted part, nor the truncated gene would produce a functional product [Ref].For illustration, we use Drosophila as a model to explain the process. A main advantage of this model organism is the availability of transposon inserts at the beginning of many genes. Classic approaches used imprecise excision of P-elements to generate deletions as described above. Here, we design sgRNAs that flank the transposon to then screen for loss of the transposon marker (e.g. GFP) after sgRNA/Cas9 mediated excision in the germline. In principle, this approach can be adapted to other model organisms and use a PCR screening approach to identify positive individuals.
For cell-lines the approach is in principle the same, but relies on chronic exposure of sgRNA/Cas9 (under selection, e.g. Puromycin) and a PCR approach for screening as detailed below.

Schematic of a typical Drosophila gene indicating the position of sgRNAs and a deletion to be generated to obtain a knock-out below the gene model. The line indicates the desired deletion and flanking dashed lines indicate positions where gRNAs can be selected. Below are primers indicated to screen for the deletion which will result in a short PCR product if the deletion is present compared to a long PCR product in the absence of a deletion, which will likely not amplify efficiently. Then, to screen for re-insertions, primers are indicated flanking an intron in the deleted part. If re-integration occurs, a short PCR product will be amplified for integration in the sense direction. If a longer PCR product is amplified, indicating a non-spliced intron, integration will be in the anti-sense direction.

Schematic of a typical human gene composed of short exons and large introns with the translated part marked in black. Removal of an exon can lead to a frameshift resulting in a truncated protein, that is non-functional (below). The line indicates the desired deletion induced by the indicated sgRNAs. RT-PCR with primers in exons flanking the deleted exon can be used to monitor CRISPR-Cas9 induced deletion efficiency in cultured cells based on the ration of splice products.
Gene insertion mutagenesis
An alternative to generate a knock-out allele is to insert a transposon into the open reading frame. Traditionally, transposon mutagenesis is random followed by selection of inserts into a given gene, but this process can be targeted by CRISPR-Cas9 genome editing to place inserts into a given gene.In Drosophila, we have generated a white gene containing PBac vector [Ref], that can be inserted in the beginning of a given gene to generate a truncated protein. Insertions in a white mutant background can then be recognized by a red eye color. If the PBac is inserted at a TTAA sequence, it can later be removed by a PBac transposase. Note, when using homology arm construct for repair, the sgRNA sites in the homology arm need to be mutated to prevent cleavage.
An extension of this approach is to include mutations in the homology arms that affect protein function. These mutations will become effective after excision of the PBac transposon.

An alternative way to make knock-outs in cultured cells, is to insert an antibiotic selection cassette in the beginning of the gene to result in a truncated protein. To ensure both alleles are knocked-out, a second insertion cassette is used, which contains a different antibiotic selection (cells are efficiently eliminated with 2.5 mg/ml neomycin, 400 µg/ml hygromycin, 0.75 µg/ml puromycin, or 4 µg/ml blasticidin S, Ishibashi et al, 2020.)

To generate conditional knock-out alleles, a degron module can be incorporated at the end of the gene (Nabet et al. 2018) again by using two selection cassettes to target both alleles.
Of note, many cell lines are aneuploid and potentially can contain more than two copies of a given gene. Also, insertion of degron cassette at the end of a gene can effect protein function.
We decline any responsibility regarding functionality of sgRNAs.
Copyright
The content including graphics and layout employed on this webpage are protected by copyright law.
Privacy | Legal | Freedom of Information | Cookies | Accessibility
