Guide for the Generation of Gene Knock-outs by Targeted Deletions or Insertions
This guide illustrates how to generate a complete gene knock-out (null allele). While single early frame-shift mutations from one sgRNA can produce hypomorphic alleles, targeted deletions or strategic insertions provide a more reliable loss-of-function outcome. (Haussmann et al.).
To begin the process, retrieve a gene model and genomic sequence for your target gene, for example from Ensembl, FlyBase, or similar databases. Protein domain annotations (PROSITE, Pfam, etc.) are shown underneath the gene model; see example here.
Gene Deletion Mutagenesis
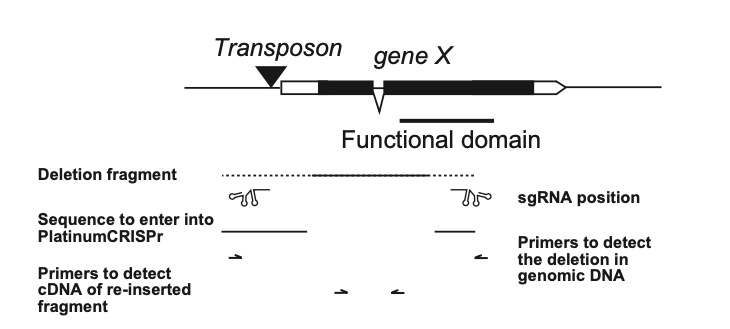
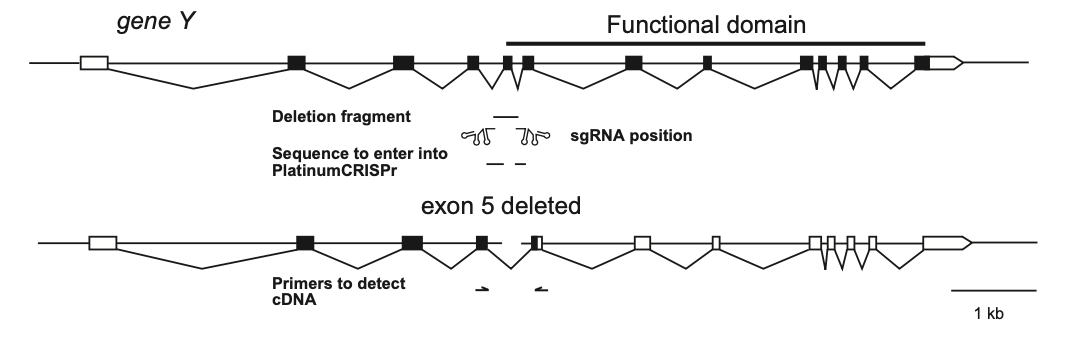
A robust way to create a null allele is to remove a portion of the promoter (for example the TATA box) together with part of an essential domain in the gene such as a catalytic domain, DNA-binding region, RNA-binding region, or similar.
Since deleted fragments can sometimes reintegrate, these re-inserted segments or truncated remnants of the gene typically do not produce functional protein products. (Haussmann et al.).
For illustration, we use Drosophila as a model system. Many fly genes contain transposon insertions near the transcription start site. Classical approaches relied on imprecise P-element excision to generate deletions. Here, CRISPR/Cas9 enables the design of sgRNAs that flank the transposon. After germline expression of sgRNA/Cas9, progeny lacking the transposon marker (e.g. GFP) can be screened to identify deletion events.
In principle, the same approach works for cultured cells: chronic exposure to sgRNA/Cas9 (with selection, e.g. puromycin) followed by PCR-based screening of the expected deletion junction.
Gene Insertion Mutagenesis
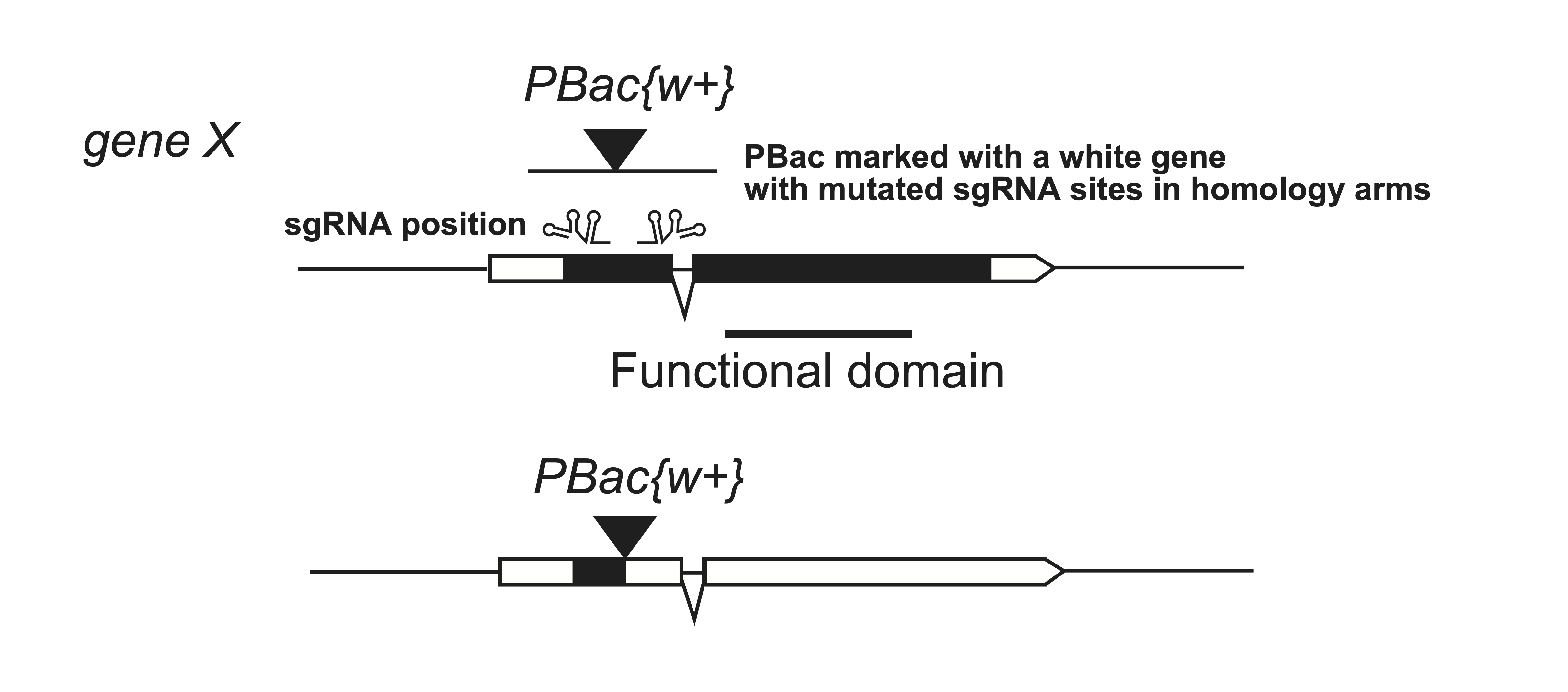
Another strategy to generate a knock-out allele is to insert a transposon within the open reading frame. Although classical transposon mutagenesis is random, CRISPR/Cas9 allows targeted placement of an insertion at a defined genomic site.
In Drosophila, a PBac-based vector carrying a white gene can be inserted at the beginning of a target gene to create a truncated protein. Successful integration yields red eye color in a white mutant background. If inserted at a TTAA motif, the PBac cassette can later be removed by PBac transposase.
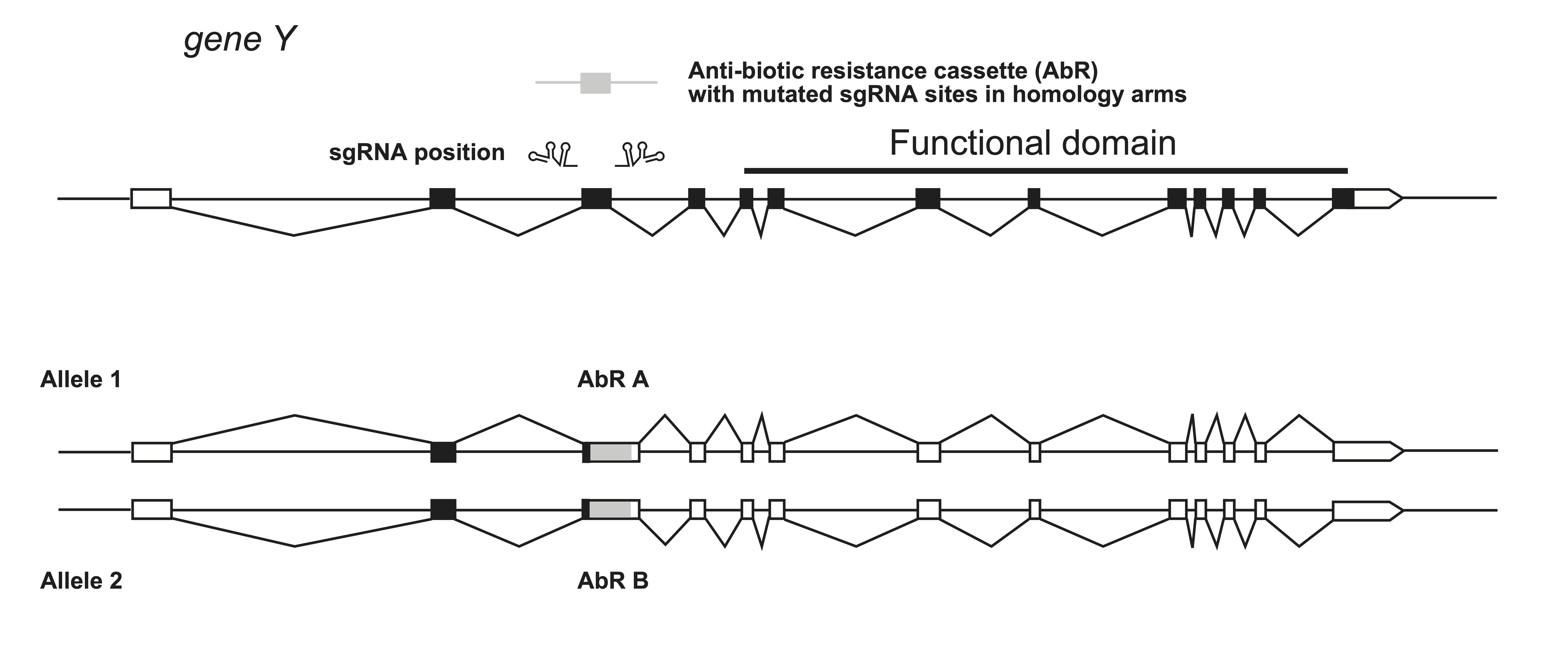
Important: When using homology-arm constructs for repair, any sgRNA target sites within the homology arms must be mutated to prevent Cas9 re-cleavage.
In cultured cells, an antibiotic selection cassette can be inserted at the start of a gene to force production of a truncated protein. To ensure complete knock-out of both alleles, a second cassette with a different selection marker is used. (Typical working concentrations: 2.5 mg/ml neomycin, 400 µg/ml hygromycin, 0.75 µg/ml puromycin, 4 µg/ml blasticidin S; Ishibashi et al., 2020 .)
To generate conditional knock-out alleles, degron modules can be added to the C-terminus of a gene (Nabet et al., 2018) . These also require two distinct selection cassettes to target both alleles.
Many cultured cell lines are aneuploid and may contain more than two gene copies. In addition, inserting a degron cassette at the end of a gene may itself influence protein function.
